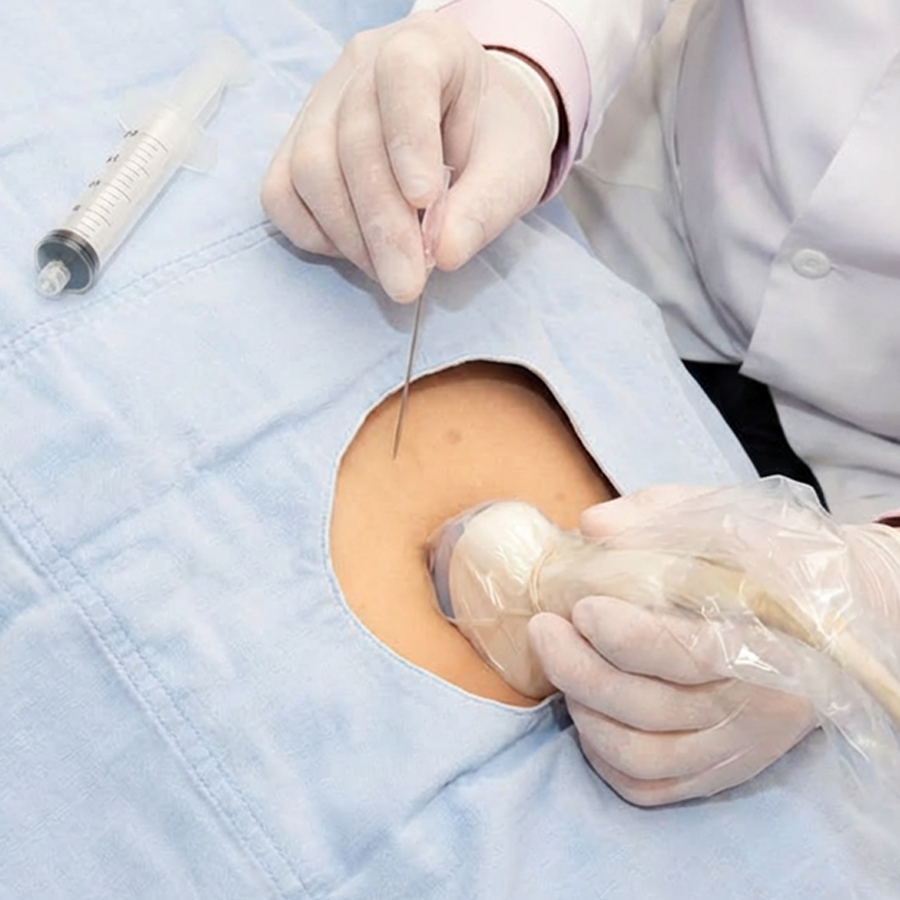
Chorionic villus sampling (CVS) and amniocentesis are ways to collect a sample, not tests in themselves.
What matters is the type of genetic test performed on the sample: PCR, FISH, karyotype, microarrays, exome, whole genome sequencing (WGS), gene-specific sequencing


Diagnostic testing (CVS or amniocentesis) examines fetal genetic material directly and can provide a definitive diagnosis for the conditions tested, rather than a risk estimate. The choice between CVS (from 11 weeks) and amniocentesis (from 15 weeks) depends on timing and the specific test required.
Increased nuchal translucency (NT) is a phenotype linked to a broad differential, many causes being rare, sporadic and de novo. To be truly diagnostic rather than merely invasive, the genetic assay must address the full differential, including monogenic disorders. A test that does not evaluate monogenic disease is not diagnostic.
Twenty years ago, karyotyping was considered diagnostic. By 2026, a truly diagnostic genomic work-up for high NT should include exome or whole-genome sequencing (WGS).
Genomics alone cannot exclude isolated structural anomalies; a targeted, expert ultrasound is still required to identify or rule these in or out.


.avif)
.avif)








QF-PCR (rapid aneuploidy screen): CVS/amnio; fast (same day–48 h) detection of T13, T18, T21 and sex-chromosome aneuploidy; can suggest triploidy.
Karyotype (G-banded): Whole-chromosome changes and large/balanced rearrangements; limited resolution; slow.
Chromosomal microarray (CMA; aCGH/SNP array): Genome-wide CNVs (~50–100 kb+); SNP arrays also assess ROH/UPD; cannot detect most single-gene variants or balanced rearrangements.
Targeted FISH: Rapid confirmation of suspected aneuploidies/rearrangements or specific microdeletions; locus-specific.
MLPA / MS-MLPA: Targeted exon-level CNVs; methylation/imprinting disorders (e.g., PWS/AS, BWS/SRS).
Single-gene testing (PCR/Sanger/NGS panels): For known familial variants or phenotype-driven monogenic disorders.
Exome sequencing (WES, ideally trio): Coding regions; SNVs/indels and some CNVs; phenotype-guided interpretation.
Whole-genome sequencing (WGS, ideally trio): Coding + non-coding; SNVs/indels/CNVs/selected SVs; best single comprehensive assay; may still miss repeats/methylation.
Low-pass (shallow) WGS: Genome-wide CNV/aneuploidy assessment; not suitable for monogenic variant detection.
Mitochondrial genome sequencing: mtDNA variants with heteroplasmy quantification; trio/segregation useful.
UPD studies (SNP array/STRs): Confirms uniparental disomy where suspected (imprinting/ROH patterns).
Quality checks: Maternal cell contamination and confined placental mosaicism assessment (STRs/array metrics); essential for result validity.
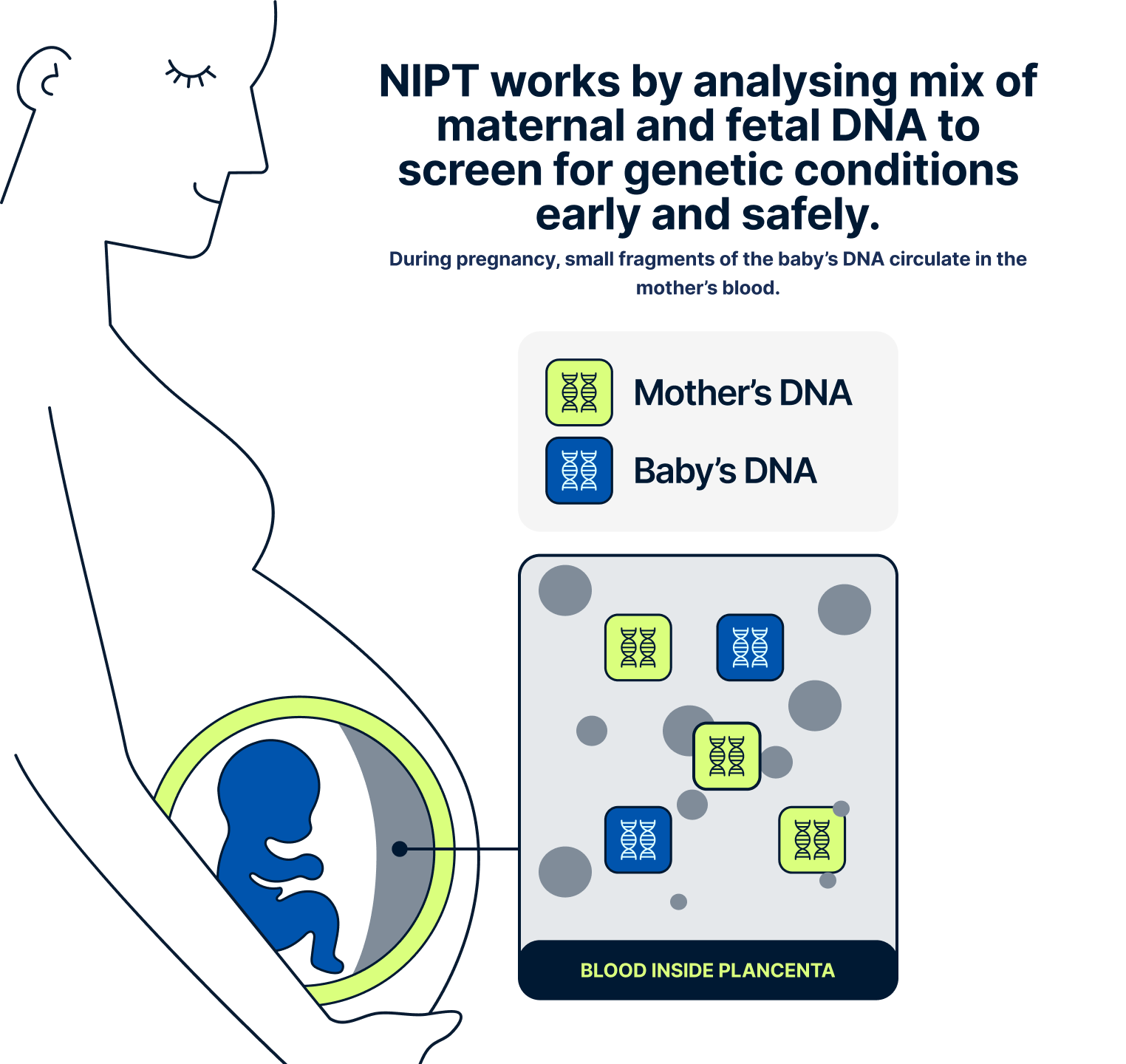
We ask you to send us your scan so our medical team can confirm that you’re at least 10 weeks pregnant and that the pregnancy appears to be developing normally. This helps us ensure the test is appropriate and that the results will be accurate. It’s also important to understand that NIPT only screens for specific genetic conditions. It does not detect structural anomalies, such as heart defects or issues with organ development, which are physical rather than genetic. These types of conditions are usually picked up later in pregnancy through detailed ultrasound scans. Sending your early scan allows our doctors to check for any concerns before moving forward with the NIPT.
Yes, NIPT can usually identify the baby’s biological sex with high accuracy, but this is completely optional. If you’d like to know, just let your genetic counsellor know during your consultation — and if you’d rather not find out, we’ll make sure it’s not included in your report.
At the moment, NIPT is only offered through the NHS in certain cases — for example, if you’ve had a high-risk result from a standard screening test. At Jeen, we offer private NIPT testing with flexible options, fast results, and full clinical support, including at-home appointments and expert guidance.
Your results will usually be ready within 2–10 working days from the time your sample arrives at our partner lab. The exact turnaround time depends on which NIPT you’ve chosen and the location of the laboratory. We’ll keep you informed throughout the process and arrange a follow-up consultation to talk you through your results as soon as they’re ready.
Please note that delays can occur during public holidays in both the UK and the US. If your results indicate that follow-up testing is needed for your partner, we’ll act quickly to keep everything moving smoothly. Our priority is to get you the answers you need without unnecessary delays — so you can make confident, informed decisions for your pregnancy.
You can take the NIPT from 10 weeks into your pregnancy. Before booking your test with Jeen, we ask you to send us a recent pregnancy scan, such as a dating or viability scan, so our team of specialists can confirm you’re eligible to proceed.
Yes, NIPT is completely safe for both you and your baby. It’s a non-invasive blood test that only requires a small sample from your arm, and there is no physical contact with the baby. This means there is no risk of miscarriage or harm to the pregnancy, unlike some invasive procedures.
NIPT is over 99% accurate at detecting Down’s syndrome and also highly reliable for Edwards’ and Patau’s syndromes. It is more accurate than standard NHS screening and produces fewer false positive results, which means fewer people are sent for unnecessary follow-up testing.
Yes, at Jeen we believe it’s important that everyone taking NIPT has a chance to speak to a qualified genetic counsellor first. This session helps you understand what the test covers, what it doesn’t, and what the results might mean for you and your baby. It’s a space to ask any questions and make confident, informed choices.
If your result shows a high risk for a condition, we’ll arrange a follow-up session with one of our genetic counsellors to explain exactly what it means. You’ll be given the option to have a diagnostic test, such as amniocentesis or CVS, which can confirm the result. We’ll support you at every step, with clear information and no pressure.
No, NIPT screens for a specific set of genetic conditions, mostly involving extra or missing chromosomes. It won’t detect all birth defects or structural anomalies, such as heart defects or limb differences. That’s why it’s important to still attend all your routine scans and check-ups during pregnancy.
Our team of experts is here to help. We're just a message away.